Introduction
I realize that, due to article length limitations on
Blogspot, in my prior blog entry,
I could not put all details about the LOINC-to-SDTM-MB for the newly publishedSARS-Cov-2 LOINC codes. So I need to provide a
few more details here before providing you the results.
LOINC code mappings for the new ARS-Cov-2 LOINC codes
These codes have been published by LOINC as a "prerelease" due to the urgency of them for use in healthcare. They are
"provisional" and regularly updated.
When starting on this project, I made the error of mapping
them to the SDTM LB domain, but soon, after some discussions with the CDISC "COVID-19 task
force" that developed the recently published Interim User Guide for
COVID-19, it became clear to me that such virology tests need to be mapped to
the MB domain (Microbiology Specimen). So
I discontinued the mapping to LB and proceeded with MB.
One of the problems with SDTM "Findings" domains
is that –TESTCD can have a pretty
different meaning depending on the domain. For LB (laboratory), LBTESTCD is the
analyte on which the measurement is being performed. Now in the case of our
COVID-19 tests, the test is either performed on Corona virus RNA, on the whole
genome, or a specific gene, or some types of antibodies. So, for
MBTESTCD/MBTEST, some new controlled terminology was necessary, which was
developed and published by CDISC by the COVID-19 task force together with the CDISC-CT team. This was then also
our first source for the mappings to MBTESTCD/MBTEST. We also made some "new term requests" for them, some making it into the interim SDTM-CT 2020-05-08,
some still under review.
An important SDTM variable then becomes MBTSTDTL ("test
detail"). Some of the possible values are "DETECTION",
"QUANTIFICATION", "VIRAL LOAD", "THRESHOLD
CYCLE". The latter is used when PCR (Polymerase Chain Reaction) is used, and the number of cycles to come to detectable (usually fluorescence)
signal is used as a measure for the amount of virus RNA: the lower the cycle
number, the higher the amount of initial virus RNA.
For MBSPEC (specimen) we tried to follow as much the
existing CDISC-CT, as we did for MBLOC (location). The interesting here is that
the "LOINC system" can either map to LBSPEC or LBLOC, as we already
also found in the LOINC-to-CDISC-LB mapping, of which the final version will
be published soon.
As usual in LOINC, "Method" is only present when
necessary to distinguish between tests when absolutely necessary, i.e. when the
results depend on the method used, or is important secondary information. This
is almost always the case for the new SARS-Cov-2 test codes. As the naming in
LOINC is pretty different from what is used in CDISC-CT, I needed to dive into
the basics of microbiology again (I studied chemistry, but that was 40 years
ago), and also has a lot of discussions with people of the CDISC COVID-19 task
force. Also here, some "new term requests" needed to be made. For example, there is no term for "rapid
immunoassay" ("IA.rapid" in LOINC), and the word
"rapid" is not to go into another SDTM-MB variable, but is still
important to be retained as it is related to the reliability of the test. So,
together with MBTESTCD/MSTEST and MBTSTDTL, the mapping to MBMETHOD was not so
easy.
Now, some of you may already have asked themselves
"where can I download the Excel with these mappings". The answer is
"you can't yet". The reason is that this mapping is still under
development, as LOINC is regularly adding new codes, and not all the necessary
CDISC-CT has been published, and, that the mapping needs further quality
control. So, only in case you are a specialist in the field, you currently can
get a copy of it with the purpose of quality control. I hope I can publish a
"final" version, once all CDISC-CT it needs is published, and after
the next formal LOINC version release, which is expected for the end of June.
The RESTful web service
You can however already use the RESTful web service that I
generated and that uses the mapping. You can find all necessary API details
here.
Please remark that the underlying database can change all the time, not only
due to additional LOINC codes being added, but also due to necessary
corrections. When requesting XML for the response, an example XML structure
that you get when querying for the LOINC code 94500-6 "SARS coronavirus 2 RNA [Presence] in
Respiratory specimen by NAA with probe detection" is:
So, you can already use the RESTful web service for
mappings, but please be aware of the limitations.
Generating SDTM-MB and DM datasets directly from FHIR
records
Of course, also the HL7-FHIR community
has picked up the COVID-19 theme, and a good number of initiatives have
started. One of the FHIR repositories that already has some of the new
SARS-Cov-2 codes in it is the "COVID19 Synth" repository of SmileCDR. It implements the highly standardized FHIR API with
the base URL being https://covid19-under-fhir.smilecdr.com/baseR4.
"Synth" stays for "synthetic", as it is contains
"synthetic" data, and is based on the famous "SyntheticMass" FHIR repository mimicking the
population of the state of Massachusetts, containing over 1 million synthetic
patient records.
So, in order to try out our mappings,
we generated a relative small Java program. It uses a lot of RESTful web
services, not only for the retrieval of records from the FHIR repository, but
also for executing the mappings (i.e. it makes calls to our own,
aforementioned, RESTful web service). The latter is very fast, the execution
time of a query is usually around 30ms. The program then generates MB and DM
SDTM datasets in the modern CDISC Dataset-XML format for all the patients in the system that have a SARS-Cov-2 record in the
repository. Also a simple define.xml was generated.
We also wanted to auto-generate an additional LB dataset for these subjects, as we demonstrated during the Virtual CDISC European Interchange last month. We found however, that for the 113 subjects found, there are no further laboratory records in the system.
We also wanted to auto-generate an additional LB dataset for these subjects, as we demonstrated during the Virtual CDISC European Interchange last month. We found however, that for the 113 subjects found, there are no further laboratory records in the system.
As we would of course like to share the
results (comments welcome!) you can download the datasets from here. As they
are in modern CDISC Dataset-XML format, you may either use e.g. the open source
"Smart Submission Dataset Viewer" or first "downgrade" them to the completely outdated SAS-XPT format
(resources can be found here).
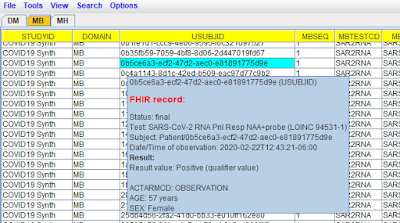
Be aware however that the MB dataset has embedded FHIR source data, which is
not possible when using SAS-XPT), and can nicely being visualized using the
"Smart Submission Dataset Viewer", like in the following example:
How to proceed?
We have now already demonstrated that
it is possible to fully automatically generate DM, LB and MB datasets from FHIR
entries in a FHIR-enabled EHR system.
I am currently also developing a LOINC-VS mapping. I identified over 600 LOINC
codes that are to be considered as a "vital sign" code. Mapping all
of these to VS is a considerable amount of work, but I am making good progress.
As soon as I get it ready, I will also make an experimental RESTful web service
for it available. Generating VS datasets directly from an EHR repository should
then be possible.
But there is other "low-hanging
fruit"! It should be pretty easily possible to generate CM (Concomitant
Medications) and MH (Medical History) SDTM datasets directly from FHIR
"MedicationAdministration" and from "Condition". Also
here, we will need to take care of different granularity (e.g.
"Condition" also covers "Risk factor") and differences in
coding systems used (HL7-FHIR often uses http://www.snomed.org/,
which is almost not used in CDISC-SDTM.
That this is really "low hanging fruit", I found out saturday (rainy) afternoon, where I could also generate the SDTM-MH dataset. For the 113 COVID-19 subjects, it contained over 28,000 records, and was generated in something like 5 minutes.
But even then, there is still a lot of work to do …