Introduction
Two things the Covid-19 pandemic made clear for clinical research is that firstly, remote clinical trials must become the standard, and secondly, that it takes too long to develop a new treatment or vaccine. For the former, already a lot is being done, such as the IMI project "Trials@Home", funded by the European Union and the European Pharmaceutical Industry (EFPIA), and to which I am a technical advisor (I acquired the project for the University of Applied Sciences FH Joanneum in Graz when I was still a professor there).
For the latter, it also becomes obvious that we need to use
much more data from electronic health records (also called "Real World
Data"), not only to be able to have many more subjects in the study (e.g.
eligibility criteria checking), but also to build a control group for comparison
of patients treated using the study drug or therapy. Especially in the times of
Covid-19, it becomes ethically questionable to have a placebo group with
subjects that do not get a treatment. Of course, this is not new, as in
oncology, where "standard treatment" is usually used for the control
group.
In the last 10 years, a lot of progress was made in using
EHRs for retrieval of clinical research data. In most cases however, this meant
that content of the CRFs was mapped to the content of the EHRs, which is very
tedious, and must be repeated for each individual study. It also does not avoid
the difficult step of mapping and converting the CRF data, even when stored in
an EDC system, to the CDISC submission SDTM standard, which is required by FDA
and PMDA for submissions to obtain a marketing authorization. After all, the
FDA and PMDA are not interested directly in the captured source data
themselves, they want to obtain the information categorized in so-called CDISC
domains datasets, and in tabular format, and with lots of derived data points.
Another caveat so far was also that EHR systems considerable differ, and an interface must be written for each EHR system.
Another caveat so far was also that EHR systems considerable differ, and an interface must be written for each EHR system.
HL7-FHIR as a game changer
All that changed when more and more EHR systems started
providing an HL7-FHIR interface "out of the
box". Some of these EHR systems are even open source, such as the
HAPI-FHIR system. One of the advantages of the HL7-FHIR
standard is that it offers a highly standardized API for use with RESTful webservices. As such, anyone with a minimal
knowledge of how to write a RESTful client, can develop and generate
applications that use data from the EHR system. Therefore, FHIR has become a
real "game changer", and thousands of applications already exist that
use the FHIR API.
Use of FHIR in clinical research
Some have seen FHIR as a serious competitor to CDISCstandards such as ODM and SDTM, but this is not correct. Information captured in FHIR resources is usually "event driven", i.e. the patient comes to the doctor or hospital with a problem, and the "care plan" is regularly adapted, depending on findings from examinations and e.g. laboratory information. In clinical research, the "plan" is governed by the study protocol, and translated into "visits", with "forms" and "questions" or "data points", mostly baked into an EDC system. Exchange of information is then usually done in CDISC ODM format. FDA and PMDA are however not interested in ODM exports, they want to have all data categorized and submitted as SDTM and after analysis, also as ADaMdatasets, and in tabular format.
FHIR however is not tabular at all, FHIR is about "linked data", in
this case about "linked resources". So, with the current preference
of the regulatory authorities to use categorized data in tabular format, it is
pretty unlikely that FDA and PMDA will one day require FHIR as a format for
submissions.
What we can however already do right now, is to deliver theFHIR source data point together with the SDTM record, as has been demonstrated several times before.
This would however require FDA and PMDA
to finally move from the outdated transport SAS-XPT format to a modern
transport format such as CDISC Dataset-XML or a JSON-based format. For SDTM, SEND and ADaM submissions in Dataset-XML
format, even an open source viewer is available that also enables to visualize
embedded source records in FHIR format.
But what if we could generate the by the regulatory required
SDTM datasets directly from a FHIR-EHR repository? This would already allow to
win considerable time for "control group" datasets. But even for
patients in a controlled clinical study, HL7 has developed a few special
resources, such as the "ResearchStudy"
and "ResearchSubject" resources. The former defines all the major characteristics of the study and also
links to a "PlanDefinition", which can be seen as the electronic
version of the study protocol. The "ResearchSubject" resource contains information about the
subject, such as the start and end of participation, reference to the
electronic confirmed consent document, the trial arm assigned to, and a patient
identifier allowing to link to all other medical information resources, such as
medications, findings, medical history.
So, essentially, if all the medical information of a
clinical research subject is in the EHR system, and can be accessed using the
FHIR API, it should in principal be possible to generate
almost all SDTM datasets fully automatically and "on the fly".
Connecting FHIR to SDTM
FHIR is essentially consisting of "resources" which can be compared to CDISC SDTM "domains". Well, sort of … Whereas some resources map pretty well 1:1 to SDTM domains, such as the Patient resource (to SDTM DM "demographics" domain), the MedicationAdministration resource (to the CM "concomitant medications" domain), the AdverseEvent resource, there are also a lot of differences.
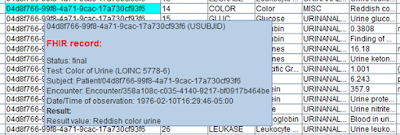
The major problem lies in mapping the FHIR Observation resource. It corresponds to the full set of SDTM "Findings" domains.
In SDTMIG v.3.3, there are 28 "Findings" domains,
for which there is only FHIR "Observation" resource. As
"Findings" datasets consist of the bulk of an electronic regulatory
submission, how can we know which "Observation" data point must go
into which SDTM domain dataset? This categorization step is already now the
most difficult step when generating SDTM datasets: the most asked question in several
of the public forums on SDTM is: "where,
in which domain (and how) do I put datapoint XYZ?". FHIR Observation
instances do not include that information. Sometimes, one can use the
"category" information, but this is not always the case, and it is
not always populated. Also, we see that in SDTM, domains are often split between
versions of the Implementation Guide, and instructions what to put where is changing. For example, COVID-19
lab test results that measure the presence or amount of viral RNA are now to be put into MB (microbiology) datasets, whereas in
the past, they were envisaged to go into LB (laboratory).
FHIR observations and LOINC codes
How does FHIR then distinguish between the type and/or
specifics of individual observations? Although there is no FHIR obligation to
use LOINC codes, almost all EHR systems differentiate
between the type of the observation by the LOINC code for the test that leads
to the observation.
LOINC is a worldwide standard for defining tests in a very exact way in healthcare and assigns a unique code to each of them. The latest version (2.67) has over 92,000 tests defined. Many of them are lab tests (including virology), but there are also many test codes for vital signs, for ECG, for standardized questionnaires, etc.. LOINC is much more specific than CDISC –TESTCD codes, the major different being that LOINC is precoordinated whereas CDISC-CT is post-coordinated. So, in order to find out to which SDTM domain a FHIR "Observation" data point belongs, one would need to map all LOINC codes to CDISC controlled terminology, i.e. for each of them decide to which SDTM domain it maps to, and then assign CDISC-CT values for –TESTCD, --TEST, --SPEC, --LOC, --LAT, --EVINT, etc..
As there are over 92,000 LOINC codes, one can already see this would be a gigantic task, to be repeated each time that a new SDTM-IG version (or even CDISC-CT version) is published, due to further "specialization" of SDTM domains with each new SDTM-IG version.
LOINC is a worldwide standard for defining tests in a very exact way in healthcare and assigns a unique code to each of them. The latest version (2.67) has over 92,000 tests defined. Many of them are lab tests (including virology), but there are also many test codes for vital signs, for ECG, for standardized questionnaires, etc.. LOINC is much more specific than CDISC –TESTCD codes, the major different being that LOINC is precoordinated whereas CDISC-CT is post-coordinated. So, in order to find out to which SDTM domain a FHIR "Observation" data point belongs, one would need to map all LOINC codes to CDISC controlled terminology, i.e. for each of them decide to which SDTM domain it maps to, and then assign CDISC-CT values for –TESTCD, --TEST, --SPEC, --LOC, --LAT, --EVINT, etc..
As there are over 92,000 LOINC codes, one can already see this would be a gigantic task, to be repeated each time that a new SDTM-IG version (or even CDISC-CT version) is published, due to further "specialization" of SDTM domains with each new SDTM-IG version.
Now, and very unfortunately, most people and organizations
in the CDISC community, still see LOINC codes as something that is a burden rather
than an opportunity, and only to be looked after in case the FDA requires the
LOINC code in a submission, as has recently become the case for the LB domain.
As a reaction on this FDA requirement, CDISC developed a mapping between the LOINC codes of the 1,400 most popular LOINC codes and the LB domain variables and controlled terminology.
This mapping already helps (limited to these 1,400 codes) to automatically generate SDTM-LB datasets from a FHIR repository for all patients in a clinical trial. This was already demonstrated during the last CDISC European Interchange.
As a reaction on this FDA requirement, CDISC developed a mapping between the LOINC codes of the 1,400 most popular LOINC codes and the LB domain variables and controlled terminology.
This mapping already helps (limited to these 1,400 codes) to automatically generate SDTM-LB datasets from a FHIR repository for all patients in a clinical trial. This was already demonstrated during the last CDISC European Interchange.
But a clinical trial submission is of course much more than
just lab data. Our investigations show that it is already easy to generate an
almost complete DM (Demographics) data set, mostly except for race and
ethnicity data which is not always present in the FHIR records, and information
about planned and actual arm. These can however in future be retrieved from the
FHIR ResearchSubject resource instances.
So, how can we extend this (limited) mapping between LOINC
codes and CDISC domains and controlled terminology? I will elucidate more about our current efforts in that field in the next blog.
Extending the mapping for SARS-Cov-2 new LOINC tests and codes
In view of the Covid-19 pandemic and crisis, LOINC
recently started developing and publishing special "prerelease" codes for tests related to SARS-CoV-2. When
discussing these with the team that generated the CDISC "Interim UserGuide for COVID-19", we became aware that these newly developed codes need
to be mapped to the MB domain, and not to the LB domain. We got a lot of help
from the team, for which we are very grateful!
Also for these codes, a RESTful web service was developed and made available. This mapping is continuously being extended, as LOINC is regularly adding new LOINC codes for SARS-CoV-2 tests.
Also for these codes, a RESTful web service was developed and made available. This mapping is continuously being extended, as LOINC is regularly adding new LOINC codes for SARS-CoV-2 tests.
In the next blog, I will show some of our results, such as
automatically generated SDTM DM, MB and LB datasets generated for 131 Covid-19
patients from the SmileCDR synthetic Covid-19 FHIR repository.
Goal of this effort to create code for generating as many SDTM datasets as
possible completely automatically from the FHIR repository. If we make good
progress, we will apply for presenting these results at the International CDISC Interchange in October.
No comments:
Post a Comment