This blog entry is an extension to the discussions on thePinnacle21 forum and a discussion on the “LinkedInSDTM Experts” forum regarding the “standardization” of “original
result units”, and the controversial CDISC
“UNIT”
codelist, and the use of UCUM notation.
We live in an era where artificial intelligence (AI) and
machine learning (ML) are booming. But, in order to apply it to CDISC SDTM, we
need an information basis. This basis should probably be the SDTM-IG.
Unfortunately, the SDTM-IGs are all published as PDF documents, which are
essentially not machine-interpretable. This makes the information very hard to
implement in software, and e.g. apply AI.
For example, when I work with electronic health records
(EHRs), for which I get the blood pressure readings with the unit “mm[Hg]” (UCUM notation – THE international standard
for unit notation), and I want to know whether I should copy “mm[Hg]” into
VSORRESU, or whether I need to replace it with the CDISC notation “mmHg” from
the CDISC “UNIT” codelist, I must (as a human) start “fighting” myself through the SDTM-IG and hope
to find an answer there.
What would such an SDTM-AI system need to be able to answer such questions? First it needs to know that “mm[Hg]” and “mmHg” are semantically identical. However, “mm[Hg]” isn’t even listed in the CDISC controlled terminology as a “synonym”, this although it is a term from the international standardized notation in international sciences, healthcare, engineering, and business. Secondly, it would require that the SDTM-IG is in a machine-readable format (PDF isn’t), with clearly defined rules (if possible machine-interpretable, like in the CDISC “SDTM-IG Conformance Rules”).
If we look at the different versions of the SDTM-IG, we
observe that the published (PDF) document grows considerably with each new version.
For example, SDTM-IG v.3.1.2 (2008) has 298 pages, whereas the version 3.2 (2013) has
398 pages, excluding the IGs for medical devices (additional 60 pages) and for
associated persons (additional 31 pages). When I first took the 2-day SDTM
course a number of years ago, the trainer (Peter VR) could explain everything
(including all domains and the “assumptions”) in these 2 days. I recently asked
him whether this is nowadays still possible and he answered me that it is not,
and that the training can only treat the principles, and even not all the
domains. Also the complexity has increased, and in my opinion, the “learning curve” has considerably steepened with each new version of the
IG, making it very hard for beginners to start working with SDTM. I sometimes
think about asking our university to set up a master degree in CDISC-SDTM, with
addition of an additional study year at each new published version of the SDTM-IG ...
At the same time, the SDTM validation software used by the FDA
(developed by a company with no ties to CDISC) has contributed to the
confusion by defining validation rules that are over-interpretations of the
SDTM-IG, are just wrong or throw a large amount of false positives.
The “SDTM-IG Conformance Rules” published by CDISC itself were a great step forward, but essentially came too late – they should have been published together with the SDTM-IG itself, and even better, as part of the IG. These rules have also been implemented under the “OpenRules for CDISC Standards”initiative in a modern, completely open format.
The “SDTM-IG Conformance Rules” published by CDISC itself were a great step forward, but essentially came too late – they should have been published together with the SDTM-IG itself, and even better, as part of the IG. These rules have also been implemented under the “OpenRules for CDISC Standards”initiative in a modern, completely open format.
SDTM and artificial intelligence (AI)
Instead of needing to have a “master
degree in SDTM”, wouldn’t it be better that one (or even
better, our computer programs) just queries something like “Alexa, can I put mm[Hg]
in VSORRESU?”? Alexa then would probably
answer something like “No,
Jozef, you need to replace mm[Hg] by mmHg as both are semantically the same and
the latter is part of CDISC controlled terminology and the former is not, and
CDISC does unfortunately not allow UCUM notation yet”.
A first simple proposal for a machine-readable SDTM-IG has been made in the past, but this proposal seems to have gone almost unnoticed by the SDTM team (SDTM team members: please correct me if I am wrong!), and the next version of the IG (likely to have 500 pages or more?) will be published as … PDF. A request to also publish the next SDTM-IG as XML has unfortunately been turned down by the CDISC SDTM team:
In order for the "SDTM-Alexa" (SDTM-AI system) to provide an answer to the question whether "mm[Hg]" (from the EHR) can be put into VSORRESU, the system needs to find the guidance in the SDTM-IG. Here it is (with many thanks to Carlo R for looking up and bringing it up in the discussion - ), from “Assumption 7” in the SDTM-IG, LB (“laboratory”) section:
essentially stating that one should first check whether the
own term/unit (in this case “mm[Hg]”) is listed as a “synonym” for “something else” in the CDISC controlled terminology, and if not found, a “new term request” should be submitted to CDISC.
Honestly said, the latter is not a real option, as this process usually takes 6 months or more, and if the request is turned down, zero progress is made.
Honestly said, the latter is not a real option, as this process usually takes 6 months or more, and if the request is turned down, zero progress is made.
In our earlier proposed prototype of a SDTM-IG in XML, each “Assumption” is an own XML element instance, for example:
Although structured, this doesn't make it machine-executable nor suitable for AI. In order to make it usable for AI or ML, we need a machine-executable expression or an algorith, which could look like:
Although structured, this doesn’t make it machine-executable. In order to be able to use
this in AI, we need a machine-executable expression or an algorithm, which
could look like:
a) Submit the “suspected synonym” to a web service (or other system) that looks whether the value (“mm[Hg]” in this case) has been published by CDISC-CT as a synonym, and if so, for what it is a synonym.
b) If the answer is “no” (or “null”), automatically make a request to the NCI
b) If the answer is “no” (or “null”), automatically make a request to the NCI
As the latter is not really an option, b) could be replaced by:
c) Extend the “UNIT” codelist in the define.xml with the own term, and put the “own term” in VSORRESU.
For step a) I created a RESTful web service this morning
which is documented at:
http://xml4pharmaserver.com/WebServices/#sdtmtermsynonym
http://xml4pharmaserver.com/WebServices/#sdtmtermsynonym
If one submits our example “mm[Hg]” to the RESTful web service (http://www.xml4pharmaserver.com:8080/CDISCCTService/rest/SDTMTermFromSynonym/mm%5BHg%5D),
one obtains:

Containing an empty response meaning that “mm[Hg]” is not found to be a “synonym” of anything. This is rather strange as this is the mandatory notation in EHRs, but this does not seem to be honored yet by the CDISC-CT team.
Another example would be e.g. that we have measured a concentration in “mol/m3” for which we than submit a request to the RESTful web service with the result:
stating that there IS a synonym for “mol/dm3” and
that we need to replace it by “mmol/L” in LBORRESU.
So, all that would be needed is that such an algorithm is
expressed as a machine-readable expression, and add it as such (probably by
using a child element) to the “Assumption” element in the XML version of the
SDTM-IG.
Some other “rules” or “assumptions” that could easily be implemented in
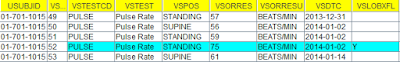
a machine-readable SDTM-IG and can then be used for AI are things like that “—DY” values
are not allowed to be “0”, so that a system could ask
questions like “Alexa, can VSDY be 0?”
This is just one of the first ideas I have for first coming
to a machine-readable SDTM-IG and then to “smart
SDTM systems” using “artificial intelligence” or “machine
learning”. This would not only greatly
help flattening the very steep (and with each IG-version becoming steeper)
learning curve, but also allow to automate mapping steps that are now done
manually and, maybe even more important, help avoiding the many different
interpretations of the SDTM-IG.
It however requires that the SDTM development team moves
away from generating the SDTM-IG from Word documents (CDISC-JIRA may be of help
here), with highly structured content (can be done using a database) and that
the team allows specialists from other domains (XML, AI, …) to work with them and have a voice
in the development.
We have self-driving cars, but for SDTM, we still rely on
30-year old technology. High time that we do something about this.